This repository hosts the code base for the paper
Automated Hypothesis Validation with Agentic Sequential Falsifications
Kexin Huang*, Ying Jin*, Ryan Li*, Michael Y. Li, Emmanuel Candès, Jure Leskovec
Link to Paper
If you find this work useful, please consider cite:
@misc{popper,
title={Automated Hypothesis Validation with Agentic Sequential Falsifications},
author={Kexin Huang and Ying Jin and Ryan Li and Michael Y. Li and Emmanuel Candès and Jure Leskovec},
year={2025},
eprint={2502.09858},
archivePrefix={arXiv}
}
Hypotheses are central to information acquisition, decision-making, and discovery. However, many real-world hypotheses are abstract, high-level statements that are difficult to validate directly. This challenge is further intensified by the rise of hypothesis generation from Large Language Models (LLMs), which are prone to hallucination and produce hypotheses in volumes that make manual validation impractical. Here we propose Popper, an agentic framework for rigorous automated validation of free-form hypotheses. Guided by Karl Popper's principle of falsification, Popper validates a hypothesis using LLM agents that design and execute falsification experiments targeting its measurable implications. A novel sequential testing framework ensures strict Type-I error control while actively gathering evidence from diverse observations, whether drawn from existing data or newly conducted procedures. We demonstrate Popper on six domains including biology, economics, and sociology. Popper delivers robust error control, high power, and scalability. Furthermore, compared to human scientists, Popper achieved comparable performance in validating complex biological hypotheses while reducing time by 10 folds, providing a scalable, rigorous solution for hypothesis validation.
We highly recommend using a virtual environment to manage the dependencies.
conda create -n popper_env python=3.10
conda activate popper_envFor direct usage of Popper, you can install the package via pip:
pip install popper_agentFor source code development, you can clone the repository and install the package:
git clone https://github.com/snap-stanford/POPPER.git
cd POPPER
pip install -r requirements.txtAdd the OpenAI/Anthropic API key to the environment variables:
export OPENAI_API_KEY="YOUR_API_KEY"
export ANTHROPIC_API_KEY="YOUR_API_KEY"Datasets will be automatically downloaded to specified data folder when you run the code.
A demo is provided in here to show how to use the Popper agent to validate a hypothesis and basic functionalities of the Popper agent.
from popper import Popper
# Initialize the Popper agent
agent = Popper(llm="claude-3-5-sonnet-20240620")
# Register data for hypothesis testing;
# for bio/discoverybench data in the paper,
# it will be automatically downloaded to your specified data_path
agent.register_data(data_path='path/to/data', loader_type='bio')
# Configure the agent with custom parameters
agent.configure(
alpha=0.1,
max_num_of_tests=5,
max_retry=3,
time_limit=2,
aggregate_test='E-value',
relevance_checker=True,
use_react_agent=True
)
# Validate a hypothesis
results = agent.validate(hypothesis="Your hypothesis here")
# Print the results
print(results)Popper supports inferencing with local LLM servers such as vLLM, SGLang, and llama.cpp, as long as they support OpenAI-compatible API. Here are some example usage with locally hosted LLMs:
Using SGLang:
# mistral large 2 with SGLang, using 4 GPUs with 8-bit quantization
python -m sglang.launch_server --model-path mistralai/Mistral-Large-Instruct-2411 --port 40000 --host 0.0.0.0 --tp 4 --quantization fp8 --mem-fraction-static 0.8 --trust-remote-codefrom popper import Popper
agent = Popper(llm="mistralai/Mistral-Large-Instruct-2411", is_locally_served=True, server_port=40000)
agent.configure(alpha=0.1)
agent.register_data(data_path='path/to/data', loader_type='bio')
agent.validate(hypothesis = 'YOUR HYPOTHESIS')Using vLLM:
vllm serve NousResearch/Meta-Llama-3-8B-Instruct --dtype auto --api-key token-abc123from popper import Popper
agent = Popper(llm="NousResearch/Meta-Llama-3-8B-Instruct", is_locally_served=True, server_port=8000, api_key="token-abc123")Using llama.cpp:
llama-server -m model.gguf --port 8080from popper import Popper
agent = Popper(llm="qwen2 1.5B", is_locally_served=True, server_port=8080)You can simply dump in a set of datasets in your domain (e.g. business, economics, political science, etc.) and run Popper on your own hypothesis. We only expect every file is in a csv or pkl format.
from popper import Popper
agent = Popper(llm="claude-3-5-sonnet-20240620")
agent.configure(alpha = 0.1)
agent.register_data(data_path='path/to/data', loader_type='custom')
agent.validate(hypothesis = 'YOUR HYPOTHESIS')You can arbitrarily define any free-form hypothesis. In the paper, we provide two types of hypothesis: biological hypothesis and discovery-bench hypothesis.
You can load the biological hypothesis with:
from popper.benchmark import gene_perturb_hypothesis
bm = gene_perturb_hypothesis(num_of_samples = samples, permuted=False, dataset = 'IL2', path = path)
example = bm.get_example(0)It will return something like:
{'prompt': 'Gene VAV1 regulates the production of Interleukin-2 (IL-2).',
'gene': 'VAV1',
'answer': 2.916,
'binary_answer': True}
num_of_samples is the number of samples you want to generate, permuted is whether you want to permute the dataset for type I error estimation, and dataset is the dataset you want to use and you can choose from IL2 and IFNG.
For discovery-bench, you can load the hypothesis with:
from popper.benchmark import discovery_bench_hypothesis
bm = discovery_bench_hypothesis(num_samples = samples, path = path)
example = bm.get_example(0)It will return something like:
{'task': 'archaeology',
'domain': 'humanities',
'metadataid': 5,
'query_id': 0,
'prompt': 'From 1700 BCE onwards, the use of hatchets and swords increased while the use of daggers decreased.',
'data_loader': <popper.utils.DiscoveryBenchDataLoader at 0x7c20793e9f70>,
'answer': True}
As each hypothesis in discoverybench has its own associated dataset, the example will return data_loader its own dataset.
Bash scripts for reproducing the paper is provided in the benchmark_scripts/run_targetval.sh for TargetVal benchmark and benchmark_scripts/run_discoverybench.sh for DiscoveryBench benchmark.
Note: the Popper agent can read or write files to your filesystem. We recommend running the benchmark scripts inside a containerized environments. We have provided a working Dockerfile and an example script to launch a Docker container and execute the scripts in benchmark_scripts/run_discoverybench_docker.sh.
To run paper benchmarks with locally-served models, you can simply passed in the extra parameters to the benchmark script, e.g.,
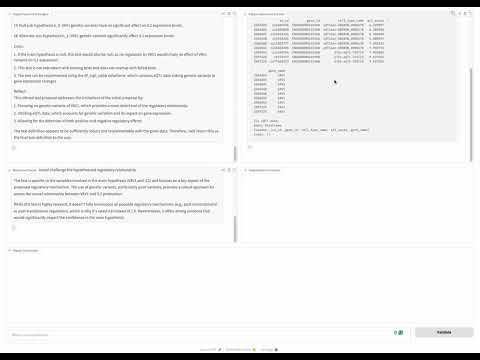
python benchmark_scripts/run_discovery_bench.py --exp_name discovery_bench --model llama-3.3-70b --num_tests 5 --samples 100 --permute --e_value --react --relevance_checker --is_locally_served --server_port 30000 --path PATH_TO_YOUR_DATASETYou can deploy a simple UI interface with one line of code using your datasets or our bio dataset - a gradio UI will be generated and you can interact with it to validate your hypothesis.
agent.launch_UI()An interface like this will be popped up:
The DiscoveryBench benchmark and some of the baseline agents are built on top of allenai/discoverybench. Thanks for their awsome work!
For any questions, please raise an issue in the GitHub or contact Kexin Huang ([email protected]).